Predicted Structures for the mtDNA peptides
Introduction:-
The coding area of the 'mtDNA' contains the genes which encode for 13 small peptides that are subsequently
used in much larger proteins involved in energy production within the cell.
The gene for each peptide are made up of a number of bases arranged in 'triplet' codons
For example: In the NAD1 gene there are 854 bases that form 318 codons.
Each codon corresponds to an amino acid and the following table shows the available amino acids.
A - Alanine (Ala) C - Cysteine (Cys) D - Aspartic Acid (Asp) E - Glutamic Acid (Glu) F - Phenylalanine (Phe) G - Glycine (Gly) H - Histidine (His) I - Isoleucine (Ile) K - Lysine (Lys) L - Leucine (Leu) M - Methionine (Met) N - Asparagine (Asn) P - Proline (Pro) Q - Glutamine (Gln) R - Arginine (Arg) S - Serine (Ser) T - Threonine (Thr) V - Valine (Val) W - Tryptophan (Trp) Y - Tyrosine (Tyr)
Non-synonymous mutations:-
A mutation in the mtDNA that leads to a change in an amino acid in a peptide is called a non-synonymous mutation.
For example:
The mutation G11778A, which is a causal mutation in Leber's Hereditary Optic Neuropathy, LHON,
changes the 340th. amino acid in the peptide NAD4 from Arginine to Histidine.
And the action of the mutation G11718A can be described as causing the non-synonymous change 'R340H' in this peptide.
Worked examples:-
The mutation G11778A leads to the non-synonymous change 'R340H' in the peptide NAD4.
This change from Arginine to Histidine will cause a change in the positive charge in part of the peptide.
The amino acids Arginine,W, Histidine,H, and Lysine,K, all carry a +ve charge
but the nature of the charge is different in each amino acid.
And by looking at the predicted structure for this peptide it is possible to see that the 340th. amino acid
of the peptide NAD4 occurs in the middle of a loop and may act as a +ve. pole.
Therefore, is is quite possible that the pathogenicity of the mutation G11718A comes about
because of the change in the nature of this +ve pole.
2nd. example:-
The mutation G15699C causes the non-synonymous change 'R318P' in the CYTB peptide and is a known pathological mutation.
The damage appears to be caused by the loss of the +ve pole when an 'Arginine' is changed to a 'Proline'.
Comparative Biology:-
For predicted structures for the peptides from various species
ATP6,
ATP8,
COX1,
COX2,
COX3,
CYTB,
NAD1,
NAD2,
NAD3,
NAD4,
NAD4L,
NAD5,
NAD6.
Human Predicted Structures:-
 Peptide ATP6; mtDNA location 8527-9204, Length 678 bases/226 codons
Peptide ATP6; mtDNA location 8527-9204, Length 678 bases/226 codons
Enlarge
 Peptide ATP8; mtDNA location 8366-8569, Length 204 bases/68 codons
Peptide ATP8; mtDNA location 8366-8569, Length 204 bases/68 codons
Enlarge
 Peptide COX1; mtDNA location 5904-7442, Length 1539 bases/513 codons
Peptide COX1; mtDNA location 5904-7442, Length 1539 bases/513 codons
Enlarge
To see a suggested 3D structure - CLICK here, then CLICK on DISPLAY
 Peptide COX2; mtDNA location 7586-8266, Length 681 bases/227 codons
Peptide COX2; mtDNA location 7586-8266, Length 681 bases/227 codons
Enlarge
To see a suggested 3D structure - CLICK here, then CLICK on DISPLAY
 Peptide COX3; mtDNA location 9207-9989, Length 783 bases/261 codons
Peptide COX3; mtDNA location 9207-9989, Length 783 bases/261 codons
Enlarge
To see a suggested 3D structure - CLICK here, then CLICK on DISPLAY
 Peptide CYTB; mtDNA location 14747-15886, Length 1140 bases/380 codons
Peptide CYTB; mtDNA location 14747-15886, Length 1140 bases/380 codons
Enlarge
To see a suggested 3D structure - CLICK here, then CLICK on DISPLAY
 Peptide NAD1; mtDNA location 3307-4260, Length 854 bases/318 codons
Peptide NAD1; mtDNA location 3307-4260, Length 854 bases/318 codons
Enlarge
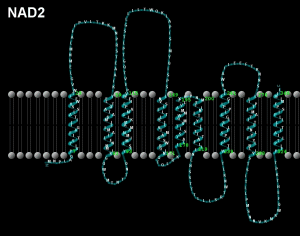 Peptide NAD2; mtDNA location 4470-5510, Length 1041 bases/347 codons
Peptide NAD2; mtDNA location 4470-5510, Length 1041 bases/347 codons
Enlarge
 Peptide NAD3; mtDNA location 10059-10403, Length 345 bases/115 codons
Peptide NAD3; mtDNA location 10059-10403, Length 345 bases/115 codons
Enlarge
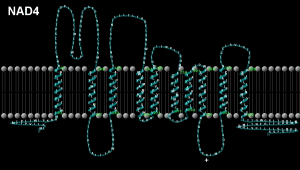 Peptide NAD4; mtDNA location 10760-12136, Length 1377 bases/459 codons
Peptide NAD4; mtDNA location 10760-12136, Length 1377 bases/459 codons
Enlarge
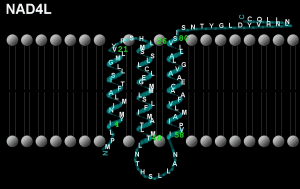 Peptide NAD4L; mtDNA location 10470-10763, Length 294 bases/98 codons
Peptide NAD4L; mtDNA location 10470-10763, Length 294 bases/98 codons
Enlarge
 Peptide NAD5; mtDNA location 12337-14145, Length 1809 bases/603 codons
Peptide NAD5; mtDNA location 12337-14145, Length 1809 bases/603 codons
Enlarge
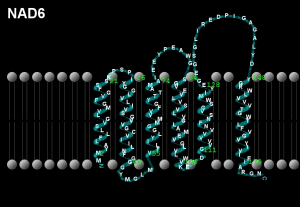 Peptide NAD6; mtDNA location 14673-14152, Length 522 bases/174 codons
Peptide NAD6; mtDNA location 14673-14152, Length 522 bases/174 codons
Enlarge
Software used:
Each diagram is produced by entering an amino acid sequence from the CRS (GenBank sequence NC_012920),
such as 'MPQLNTTVWPTMITPMLLTLFLITQLKMLNTNYHLPPSPKPMKMKNYNKPWEPKWTKICSLHSLPPQS' for the gene ATP8,
into the TMHMM Server v. 2.0 at http://www.cbs.dtu.dk/services/TMHMM/
The output from this program was then entered into TMRPres2d,
a free download from http://biophysics.biol.uoa.gr/TMRPres2D/download.jsp,
to produce the above diagrams.
If anyone knows how to produce more detailed diagrams I will be pleased to hear from you.
IF YOU HAVE ANY QUESTIONS - CONTACT ME ON: ianlogan22@btinternet.com